How to...
-
How to list all running processes in PBS-PRO
qstat -rn1tw
-
How to find a best blast hit:
-
-
How to calibrate an XYZ Printing da Vinci 2.0 A Duo 3D-printer:
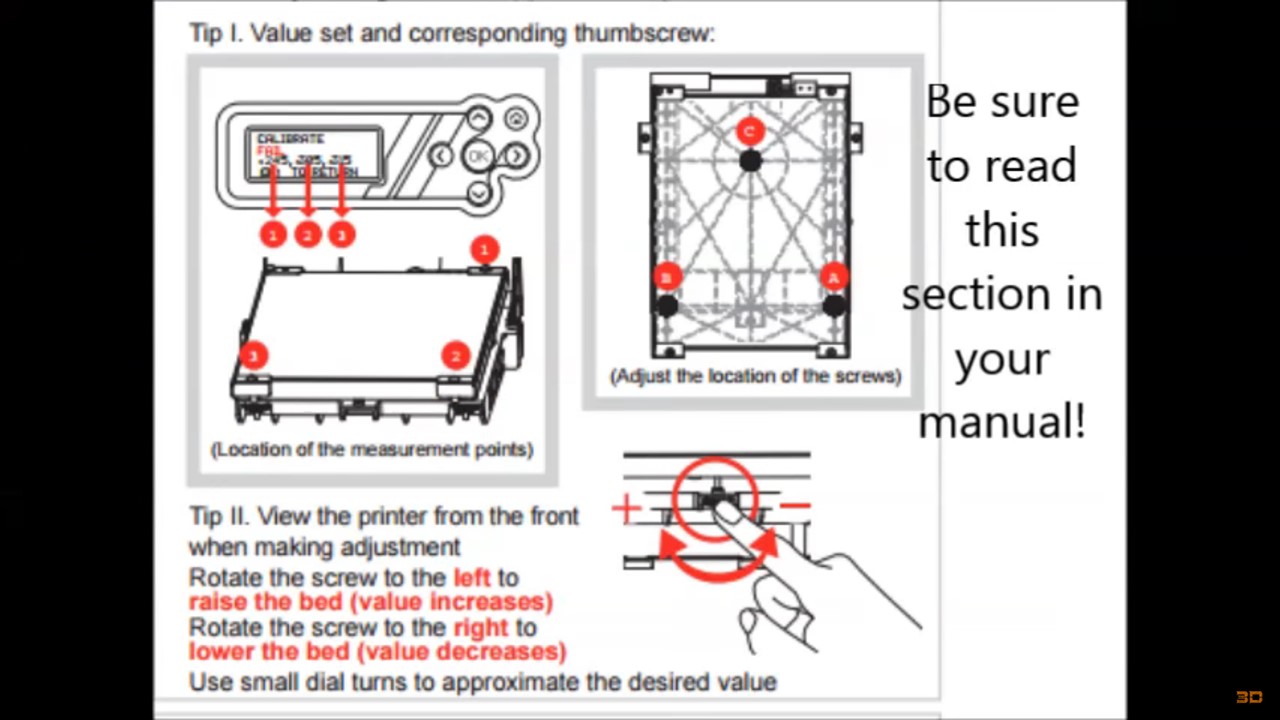
-
How to make the plastic stick to the base when 3d printing:
-
How to install point-e on Windows 11:
#in Windows Powershell note down the cuda version from this command: nvidia-smi.exe #look up the channel for your cuda version using this site: #https://anaconda.org/nvidia/repo/installers? #In Anaconda powershell: conda update conda conda update anaconda conda update python conda update --all conda create --name point-e pip #Now exit from the Powershell you are using and then open a new one before you activate the new "env". conda activate point-e #install cuda from the channel identified previously conda install cuda --channel "nvidia/label/cuda-12.0" #NB. there doesn't seem to be an option (yet) to use cud=12.0 in the code line below. The line below is suggested when visiting https://pytorch.org/ conda install pytorch torchvision torchaudio pytorch-cuda=11.6 -c pytorch -c nvidia #NB. I found that the line below was necessary to get things to work conda install cudatoolkit conda install ipykernel jupyter nvcc --version conda install -c anaconda git git clone https://github.com/openai/point-e.git cd point-e pip install -e . jupyter notebook #navigate to the notebook in Jupyter at: #./point-e/point_e/examples/text2pointcloud.ipynb #In the notebook add a cell at the beginning with the code below to test if the GPU is detectable: import torch print(f'PyTorch version: {torch.__version__}') print('*'*10) print(f'_CUDA version: ') !nvcc --version print('*'*10) print(f'CUDNN version: {torch.backends.cudnn.version()}') print(f'Available GPU devices: {torch.cuda.device_count()}') print(f'Device Name: {torch.cuda.get_device_name()}') #NB. I had an issue with the kernel crashing when plotting the pointclouds. #The error in the anaconda powershell was: #OMP: Error #15: Initializing libiomp5md.dll, but found libiomp5md.dll already initialized. #The solution was to add a cell at the start of the script with the code: import os os.environ["KMP_DUPLICATE_LIB_OK"]="TRUE" #NB. The above workaround apparently may cause crashes or silently produce incorrect results....
-
How to visualise massive point clouds in python:
https://towardsdatascience.com/guide-to-real-time-visualisation-of-massive-3d-point-clouds-in-python-ea6f00241ee0
-
How to create threaded replies in NodeBB:
-
How to negotiate a better home loan rate with your bank (from Barefoot Investor SCOTT PAPE):
You: Hello, my account number is ______. I’ve been with you for ___ years, but I’ve applied to refinance with UBank. Their rate is ____ per cent, which is a full ___ per cent cheaper than you’re charging me. Given our longstanding relationship, I’d like you to match the offer — or send me the forms I need to switch to UBank.
Bank rep: One moment, please.
(You’re bluffing, of course. However, the bank’s sales team have strict targets, backed by incentives, that they have to meet — one of which is giving profitable customers discounts to stop them leaving.)
Bank rep: We can’t match the rate you have quoted. However, we understand you are a valuable customer, so we would like to offer you a 0.15 per cent discount.
You: That’s not good enough. I’ve already got conditional approval … so in order to stay I need at least a 0.5 per cent discount. Could you please speak to your supervisor? I’m happy to wait.
Bank rep (a full six minutes later): On reviewing your case, we can offer you that 0.5 per cent discount on your current rate.
You: Brilliant! Please send me an email confirming the new rate and confirming that it will be applied as of start of business tomorrow.
-
How to fix the error "FATAL: container creation failed:" "error: can't mount image " "failed to mount squashfs filesystem: input/output error":
#This occurs when you try to run a singularity container within a VM and it is located on a mounted ntfs filesystem. The solution is to transfer the container to a different filesystem eg. ext3.
-
How to run docker-compose when you get the errors:
- File "posixpath.py", line 376, in abspath
- FileNotFoundError: [Errno 2] No such file or directory
Open a new terminal and repeat the command.
-
How to login to root account in a new ubuntu/xubuntu virtual machine that has been set up with a non-administrator account:
su yourpasswordfortheuseraccount
-
How to update apt and install filezilla on xubuntu using a proxy:
sudo apt -o Acquire::http::proxy="http://user:password@host:port/" update sudo apt -o Acquire::http::proxy="http://user:password@host:port/" install filezilla
-
How to install ubuntu on windows:
- Follow the instructions at:
https://s1gr1d.medium.com/how-to-set-up-linux-on-windows-with-wsl-2-debe2a64d20d
(NB. these instructions work for Windows 10 and 11 although the location of the Settings menus may differ slightly. Also note that VMX is the same as VT-X in the BIOS settings) - In Windows Powershell, you may also need to issue the following commands:
bcdedit /set hypervisorlaunchtype Auto wsl --update
- Follow the instructions at:
-
How to use your phone as a webcam:
-
How to fix the following error in NextFlow:
WARN: Failed to render DAG file:
This error prevents NextFlow from drawing the DAG for the pipeline.
#install Graphviz #eg. with conda conda install -c conda-forge graphviz
-
How to gather and group all tuple elements in a NextFlow channel by the first and second indexes of each tuple, perform a process on a file containing the first and second indices, and then reform the original channel by scattering:
hc_ch = Channel.of( ['abc', 1, file('file1.txt')], ['abc', 1, file('file2.txt')], ['abc', 1, file('file3.txt')], ['def', 2, file('file4.txt')] ) combinedChannel = hc_ch.groupTuple(by: [0, 1])- Expected structure of combinedChannel:
[ ['abc', 1, [file('file1.txt'), file('file2.txt'), file('file3.txt')]],
['def', 2, [file('file4.txt')]]
]process delete_txt { input: tuple val(id), val(sub_id), val(files) from combinedChannel output: tuple val(id), val(sub_id), val(files) into processedChannel script: """ rm ${id}_${sub_id}.txt """ }- Expected structure of processedChannel:
[ ['abc', 1, [file('file1.txt'), file('file2.txt'), file('file3.txt')]],
['def', 2, [file('file4.txt')]]
]flattenedChannel = processedChannel.flatMap { id, sub_id, files -> files.collect { [id, sub_id, it] } }- Expected structure of flattenedChannel:
[ ['abc', 1, file('file1.txt')],
['abc', 1, file('file2.txt')],
['abc', 1, file('file3.txt')],
['def', 2, file('file4.txt')]
]
-
How to get the path for the folder containing a bash script within the bash script:
# Get the directory where the script resides SCRIPT_DIR="$(cd "$(dirname "$0")" && pwd)" echo $SCRIPTDIR
-
How to convert a minimap2 alignment to gff/gtf:
https://github.com/lh3/minimap2/issues/455
https://github.com/lh3/minimap2/files/9591008/bam2gff_fixGffread.zip
bam2gff_fixGffread.zipminimap2 -t 10 -ax splice:hq -uf ref.fa cdna.fa |/Bio/bin/samtools-1.14 view -b > minimap2.tr.bam perl bam2gff.pl -b minimap2.tr.bam -o minimap2.tr.gff -s /Bio/bin/samtools-1.14 gffread minimap2.tr.gff -T -o minimap2.tr.gtf perl fixGffread.pl -i minimap2.tr.gtf -o minimap2.tr.fix.gtfAlternative method:
#Align sequences and convert to BAM minimap2 -ax splice --cs target.fa query.fa | samtools sort -O BAM - > alignments.bam #Convert to BED12 using BEDtools bedtools bamtobed -bed12 -i alignments.bam > alignments.bed #Convert to genePred using UCSC tools bedToGenePred alignments.bed alignments.genepred #Convert to GTF2 using UCSC tools. genePredToGtf has additional options that might be useful in specific use cases. genePredToGtf "file" alignments.genepred alignments.gtf
-
How to do a dotplot with minimap2:
minimap2 -DP ref.fa query.fa|miniasm/minidot - > dot.eps